
An End-to-End solution for “Hit to lead” towards preclinical trials
A tailored Drug design application: The application identifies leading drug candidates among the sequence space of 2 Billion of peptides, proteins or antibodies. Reducing the necessary time of the drug discovery process towards “Lead candidates” from 6 to 12 months minimum, with significant cost saving and better accuracy in results.
AI-team@peaccel.com or hit-to-lead-team@peaccel.com

Identification of smart libraries: The screening platform allows to identify not one “Hit to lead” but a multitude of drug candidates e.g. 12 million variants of a protein lead to 86,000 potential lead candidates. Increasing the chances of finding the drug candidate with the right multiple properties and reducing the risk of lead molecule failure during clinical phases (only 1/1000 molecules currently succeed in clinical trials phases).
hit-to-lead-team@peaccel.com

Don’t miss the Lead: The introduction of one or more mutations in a protein sequence can have synergistic or antagonistic effects on the protein function: This non-additivity is called epistasis and not being able to predict them can lead to missing the “Hit to Lead” variant. Our application captures epistatic effects and reduces the risk of missing a true “Hit to Lead”.
protein-team@peaccel.com
Avoiding aggregation during preclinical and clinical phases: our technology allows selection of potential Lead candidates that will not be affected by protein aggregation. Aggregation of peptides, proteins and antibodies during clinical trials phases is a major cause of failure during drug development.
hit-to-lead-team@peaccel.com

Optimization of FDCs: Our platform combines existing already marketed drugs, hence where Proof Of Concept has already been demonstrated, clinical stages have been successfully cleared and FDA approved. Optimized combinations are identified: Fixed-Dose Combination Drugs Market is up to $9bn.
FDCs-team@peaccel.com
Identifying Drug targets through dedicated AI tools
Combined tailored AI approaches for Biomarkers: Metabolic pathway modeling plays an increasing role in drug design by allowing better understanding of the underlying regulation and controlling networks in the metabolism. Our tailored Data driven approaches combined to constraint modelling allow easier and more precise drug target identification.
metabolic-pathways-team@peaccel.com or drug-target-team@peaccel.com

Identification of smart libraries: The screening platform allows to identify not one “Hit to lead” but a multitude of drug candidates e.g. 12 million variants of a protein lead to 86,000 potential lead candidates. Increasing the chances of finding the drug candidate with the right multiple properties and reducing the risk of lead molecule failure during clinical phases (only 1/1000 molecules currently succeed in clinical trials phases).
hit-to-lead-team@peaccel.com

Testing a full range of ML and DL modelling approaches: Choosing a modeling approach a priori can be counterproductive: it may not be adapted to the problem to be solved. Depending on the question asked, our application proposes and tests different ML and DL approaches to finally choose the most suitable one for your problem.
AI-team@peaccel.com
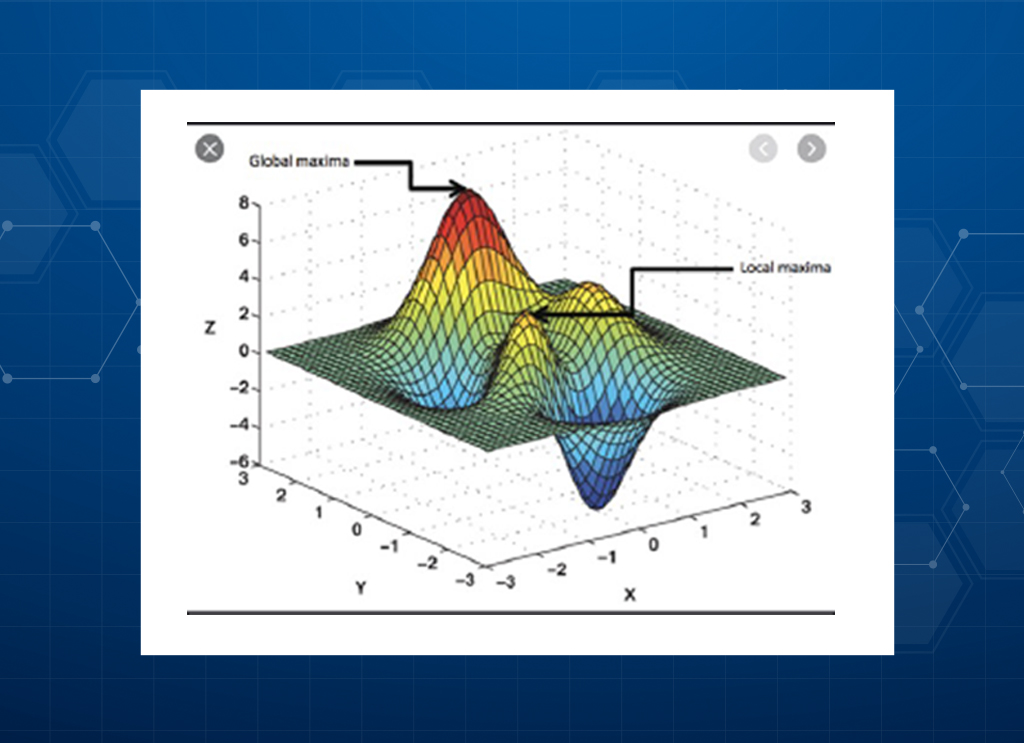
Don’t miss the real Lead!
Protein optimization problem: The solution identifies the best combination of mutations to improve desired property or function of a protein (enzymes, antibodies, peptides, etc.). Benefits include reduced costs, time and risks associated with the engineering of proteins and metabolic pathways: e.g. improvements up to 120-folds are obtained and epistatic effects captured.
protein-team@peaccel.com

Don’t miss the Lead: The introduction of one or more mutations in a protein sequence can have synergistic or antagonistic effects on the protein function: This non-additivity is called epistasis and not being able to predict them can lead to missing the “Hit to Lead” variant. Our application captures epistatic effects and reduces the risk of missing a true “Hit to Lead”.
protein-team@peaccel.com

A tailored screening
application: The application screens candidates among the sequence space of 1 billion peptides, proteins or antibodies in one day. Reducing the necessary time towards “Lead candidates” from 6 to 12 months minimum, with significant cost savings and better accuracy in results.
AI-team@peaccel.com

Reducing the risks of patent loopholes
Protection of a family of biologics : The combination of all interesting mutation positions of a protein sequence is achieved. The combinations that improve the performance of the molecules are all identified and protected, thus reducing the risks of patent loopholes and the emergence of competing molecules.
AI-team@peaccel.com

Optimal Enzymes or Bioreactors for higher yields
Bioreactors with higher performance: One of the challenges in a cell-free system is selecting the optimized enzyme concentration for optimal yield. Our customized Machine Learning Approach allows efficient Selection of enzyme concentrations and its application for Flux Optimization: e.g. flux improvements of up to 63% coupled with a cost decrease of up to 25% drastically reduce the production costs.
bioreactors-team@peaccel.com

Application to optimize multiple protein parameters: The application improves and changes different properties of protein across different environments. The optimal combination of mutations is determined, leading to a “Hit to lead” with better performance: controlling the trade-off between different properties allows to increase the production yield.
protein-team@peaccel.com

Application to optimize multiple enzyme parameters: The application was applied to improve an enzyme in the presence of different mediators across a range of pH values. The optimal combination of mutations was determined, leading to a mutant with 12-fold improvement in specificity for one mediator and 8-fold improvement in specificity for another mediator, compared to the wild-type enzyme, and better performance in three pH-adjusted buffers.
protein-team@peaccel.com

Application to optimize an enzyme: The application finds the optimal combination from 9 single point mutations, totaling in 512 mutants. By optimizing the combinations, the resulting improved protein showed a 50-fold improvement in enantioselectivity.
protein-team@peaccel.com
